First report of microalgae Rhexinema paucicellulare (Ulvophyceae) in Mauritius and its biochemical evaluation as a source of fatty acids
Accepted: January 13, 2022
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Authors
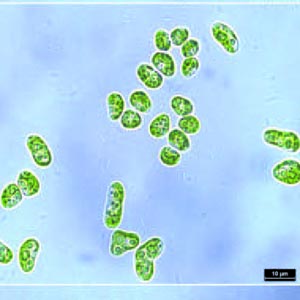
Mauritius is a tropical island with a very rich biodiversity of endemic organisms with an unexplored pool of microalgae, especially in the Ulvophyceae class. From freshwater sources, an isolate was characterised using morphological traits and 18S rDNA region. It was determined to be Rhexinema paucicellulare and was further evaluated for its biochemical content against commercially available Spirulina sp. This study evaluates the lipids content and offers a first report on the carbohydrate and protein contents of this species. The biochemical profile of Spirulina sp. differed significantly from R. paucicellulare with respect to carbohydrate (17.36% vs. 20.31%); protein (45.09% vs. 25.71%) and lipids (5.86% vs. 13.88%). Further exploration of the fatty acids profile through GC analysis revealed high presence of alpha-linolenic acid at 26.48% and linoleic acid at 6.31%. The presence of important omega-3 FA was evidenced through GC/MS analysis with eicosapentaenoic acid and docosahexaenoic acid, which could make this isolate a potential candidate in the field of aquaculture.
How to Cite
PAGEPress has chosen to apply the Creative Commons Attribution NonCommercial 4.0 International License (CC BY-NC 4.0) to all manuscripts to be published.

 https://doi.org/10.4081/jbr.2022.9950
https://doi.org/10.4081/jbr.2022.9950



