Absence of mutations at SERPINI1 gene in a cohort of patients with Cerebral Cavernous Malformations
Accepted: August 21, 2021
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Authors
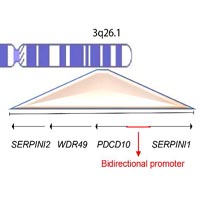
Cerebral Cavernous Malformations (CCM) are vascular lesions affecting brain microvessels. While molecular bases of the sporadic condition are not yet well elucidated, familial forms arise following mutations at three different loci KRIT1, CCM2 and PDCD10. However, no germline mutations are detected in a small percentage of families with hereditary history of CCM. In order to detect other possible candidate genes, we performed molecular analysis of SERPINI1 gene in a cohort of patients carrying no mutations in the three CCM loci, aiming to detect mutations likely associated to lesion development. Therefore, we performed molecular analysis of the SERPINI1 gene in a cohort of 18 unrelated patients affected by both familial and sporadic CCM showing no germline causative mutations. Mutational analysis resulted negative and only few single nucleotide polymorphisms were detected. However, the rs11284733 SNP was detected in a high percentage of patients affected by familial form of the disease. This SNP occurs within a noncoding exon retained in an alternative spliced SERPINI1 transcript, suggesting its possible role in gene expression regulation.
1Department of Biomedical and Dental Science and of Morphological and Functional Images, University of Messina, Messina, Italy
2Department of Biomolecular Strategies, Genetics, Vanguard Therapies and Neuroscience, I.E.ME.S.T., Palermo, Italy
How to Cite
PAGEPress has chosen to apply the Creative Commons Attribution NonCommercial 4.0 International License (CC BY-NC 4.0) to all manuscripts to be published.

 https://doi.org/10.4081/jbr.2021.9838
https://doi.org/10.4081/jbr.2021.9838



