Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.
Proteomic profiling of the brain from the wobbler mouse model of amyotrophic lateral sclerosis reveals elevated levels of the astrogliosis marker glial fibrillary acidic protein
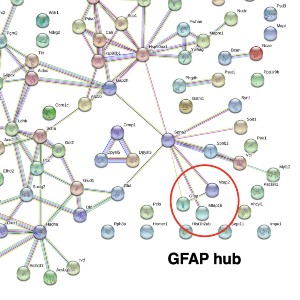
The wobbler mouse is a widely used model system of amyotrophic lateral sclerosis and exhibits progressive neurodegeneration and neuroinflammation in association with skeletal muscle wasting. This study has used wobbler brain preparations for the systematic and mass spectrometric determination of proteome-wide changes. The proteomic characterization of total protein extracts from wobbler specimens was carried out with the help of an Orbitrap mass spectrometer and revealed elevated levels of glia cell marker proteins, i.e., glial fibrillary acidic protein and the actin-binding protein coronin. In contrast, the abundance of the actin-binding protein neurabin and the scaffolding protein named piccolo of the presynaptic cytomatrix were shown to be reduced. The increased abundance of glial fibrillary acidic protein, which is frequently used in neuropathological studies as a marker protein of glial scar formation, was confirmed by immunoblotting. In analogy, the proteomic profiling of the brain from another established murine model of motor neuron disease, the SOD1mouse, also showed increased levels of this intermediate filament protein. This suggests that neurodegenerative processes are associated with astrogliosis in both the wobbler and SOD1 brain.
How to Cite

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
PAGEPress has chosen to apply the Creative Commons Attribution NonCommercial 4.0 International License (CC BY-NC 4.0) to all manuscripts to be published.

 https://doi.org/10.4081/ejtm.2023.11555
https://doi.org/10.4081/ejtm.2023.11555