Mass spectrometry-based proteomic characterization of the middle-aged mouse brain for animal model research of neuromuscular diseases
Accepted: 28 July 2023
HTML: 102
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Authors
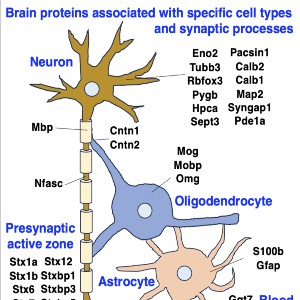
Neuromuscular diseases with primary muscle wasting symptoms may also display multi-systemic changes in the body and exhibit secondary pathophysiological alterations in various non-muscle tissues. In some cases, this includes proteome-wide alterations and/or adaptations in the central nervous system. Thus, in order to provide an improved bioanalytical basis for the comprehensive evaluation of animal models that are routinely used in muscle research, this report describes the mass spectrometry-based proteomic characterization of the mouse brain. Crude tissue extracts were examined by bottom-up proteomics and detected 4558 distinct protein species. The detailed analysis of the brain proteome revealed the presence of abundant cellular proteoforms in the neuronal cytoskeleton, as well as various brain region enriched proteins, including markers of the cerebral cortex, cerebellum, hippocampus and the olfactory bulb. Neuroproteomic markers of specific cell types in the brain were identified in association with various types of neurons and glia cells. Markers of subcellular structures were established for the plasmalemma, nucleus, endoplasmic reticulum, mitochondria and other crucial organelles, as well as synaptic components that are involved in presynaptic vesicle docking, neurotransmitter release and synapse remodelling.
How to Cite

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
PAGEPress has chosen to apply the Creative Commons Attribution NonCommercial 4.0 International License (CC BY-NC 4.0) to all manuscripts to be published.

 https://doi.org/10.4081/ejtm.2023.11553
https://doi.org/10.4081/ejtm.2023.11553



