Involvement of muscle satellite cell dysfunction in neuromuscular disorders: Expanding the portfolio of satellite cell-opathies
Accepted: 11 February 2022
HTML: 93
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Authors
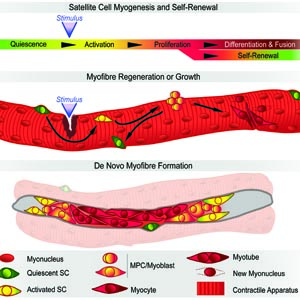
Neuromuscular disorders are a heterogeneous group of acquired or hereditary conditions that affect striated muscle function. The resulting decrease in muscle strength and motility irreversibly impacts quality of life. In addition to directly affecting skeletal muscle, pathogenesis can also arise from dysfunctional crosstalk between nerves and muscles, and may include cardiac impairment. Muscular weakness is often progressive and paralleled by continuous decline in the ability of skeletal muscle to functionally adapt and regenerate. Normally, the skeletal muscle resident stem cells, named satellite cells, ensure tissue homeostasis by providing myoblasts for growth, maintenance, repair and regeneration. We recently defined 'Satellite Cell-opathies’ as those inherited neuromuscular conditions presenting satellite cell dysfunction in muscular dystrophies and myopathies (doi:10.1016/j.yexcr.2021.112906). Here, we expand the portfolio of Satellite Cell-opathies by evaluating the potential impairment of satellite cell function across all 16 categories of neuromuscular disorders, including those with mainly neurogenic and cardiac involvement. We explore the expression dynamics of myopathogenes, genes whose mutation leads to skeletal muscle pathogenesis, using transcriptomic analysis. This revealed that 45% of myopathogenes are differentially expressed during early satellite cell activation (0 - 5 hours). Of these 271 myopathogenes, 83 respond to Pax7, a master regulator of satellite cells. Our analysis suggests possible perturbation of satellite cell function in many neuromuscular disorders across all categories, including those where skeletal muscle pathology is not predominant. This characterisation further aids understanding of pathomechanisms and informs on development of prognostic and diagnostic tools, and ultimately, new therapeutics.
How to Cite
PAGEPress has chosen to apply the Creative Commons Attribution NonCommercial 4.0 International License (CC BY-NC 4.0) to all manuscripts to be published.

 https://doi.org/10.4081/ejtm.2022.10064
https://doi.org/10.4081/ejtm.2022.10064



